Multimedia Gallery
Using biophysical protein models to map genetic variation to phenotypes
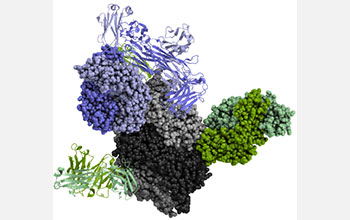
In this figure, the neutralization of infection from respiratory syncytial virus depends on antibodies (blue, green) binding to viral proteins (gray).
More about this image
The U.S. National Science Foundation made eight awards totaling $41.7 million for projects aimed at building capacity to research a national priority area: understanding the relationship in organisms between genetic material, or genotype, and physical characteristics due to gene expression and environmental influences, or phenotype.
The genotype-to-phenotype relationship has significant societal and economic implications across scientific fields and areas of industry, including but not limited to medicine, agriculture, biotechnology and ecology. An enhanced understanding of this relationship holds the potential for improved food crop yields, better prediction of human disease risk and new drug therapies. Through these investments, NSF aims to provide the scientific community with new tools and resources for future discoveries. The awards were made through NSF's Established Program to Stimulate Competitive Research as part of its Research Infrastructure Improvement Track-2 investment strategy.
"Over the past several decades, scientists and engineers have made massive strides in decoding, amassing and storing genomic data," said Denise Barnes, Head of NSF's EPSCoR. "But understanding how genomics influence phenotype remains one of the more profound challenges in science. These awards lay the groundwork for closing some of the biggest gaps in biological knowledge and developing interdisciplinary teams needed to address the challenges."
Among the awards made was for "Using biophysical protein models to map genetic variation to phenotypes," principal investigator Frederick Ytreberg, University of Idaho.
One of the great challenges in modern biology is understanding how changes in amino acids that are the building blocks of proteins lead to changes in the characteristics of a living organism. This project tackles this aspect of the genotype-phenotype challenge by focusing on protein biophysical models and using a diverse set of experimental systems, molecular and mathematical modeling. The project builds upon complementary expertise and infrastructure in Idaho, Vermont and Rhode Island. The research has significant societal implications for its potential to lead to advances in biotechnology and health sciences.
[Research supported by NSF grant OIA 1736253.]
To learn more, see the NSF News Release NSF EPSCoR awards new projects to help understand connections between genes and organisms' characteristics. (Date image taken: February 2017; date originally posted to NSF Multimedia Gallery: Dec. 13, 2017)
Credit: F. Marty Ytreberg, University of Idaho
Images and other media in the National Science Foundation Multimedia Gallery are available for use in print and electronic material by NSF employees, members of the media, university staff, teachers and the general public. All media in the gallery are intended for personal, educational and nonprofit/non-commercial use only.
Images credited to the National Science Foundation, a federal agency, are in the public domain. The images were created by employees of the United States Government as part of their official duties or prepared by contractors as "works for hire" for NSF. You may freely use NSF-credited images and, at your discretion, credit NSF with a "Courtesy: National Science Foundation" notation.
Additional information about general usage can be found in Conditions.
Also Available:
Download the high-resolution JPG version of the image. (145.4 KB)
Use your mouse to right-click (Mac users may need to Ctrl-click) the link above and choose the option that will save the file or target to your computer.